rpact in Practical Use
December 10, 2025
rpact in Practical Use

Example illustrating the package concept
Usage inspired by the typical workflow in trial design and conduct:
- Everything is starting with a design, e.g.:
design <- getDesignGroupSequential() - Find the optimal design parameters:
R / rpact - Calculate the required sample size and power, e.g.:
getSampleSizeMeans(),getPowerMeans() - Optionally perform simulations to confirm the operating characteristics, e.g.:
getSimulationMeans() - Collect your data, import it into R and create an rpact dataset:
data <- getDataset() - Analyze your data:
getAnalysisResults(design, data)
Example: Planning and Conducting a Clinical Study Using R and rpact
- In this fictive example, we will illustrate the planning and conduct of a clinical trial to assess the effectiveness of a new antihypertensive therapy.
- Each step will be demonstrated using R, primarily with the rpact package.
- Additional R packages may be used as needed.
Study Design
Objective: To evaluate the efficacy of a new antihypertensive therapy
(compared to a placebo) in patients with hypertension.
Endpoints and Assumptions:
- Endpoint: Continuous endpoint (blood pressure reduction).
- Comparison Groups: New Treatment vs. Placebo.
- Statistical Parameters:
- One-sided test with \(\alpha\) = 0.025.
- Power = 80%.
Initial Assumptions and Parameter Estimation
Assumptions:
- Patients in this study suffer from high blood pressure (grade 2 hypertension), often around 150 mmHg (systolic).
- The objective is to significantly reduce blood pressure in the treatment group, while no substantial reduction is expected in the placebo group.
Expected Values:
- Target difference in systolic blood pressure change from baseline:
12 mmHg (difference in mean reduction between treatment and placebo). - Variability of change from baseline:
Standard deviation of 15 mmHg.
Step 1: Sample Size Calculation
Using rpact we will calculate the required sample size to achieve a power of 80% for detecting a 12 mmHg difference in systolic blood pressure change from baseline in the treatment group relative to the placebo group, assuming a standard deviation of 15 mmHg.
We will see a nice feature when using rpact output in RMarkdown or Quarto docs: you directly get a nicely formatted result output.
Step 1: Sample Size Calculation (cont’d)
# Load the rpact package
library(rpact)
# Define the design parameters for sample size calculation
design <- getDesignGroupSequential(
kMax = 1, # Only one analysis (classic fixed design)
alpha = 0.025, # Significance level
beta = 0.20, # 80% power
sided = 1 # One-sided test
)
# Estimate sample size
sampleSizeResult <- design |>
getSampleSizeMeans(
groups = 2, # Two groups: Treatment vs. Placebo
alternative = 12, # Expected effect size
stDev = 15 # Common standard deviation
)Step 1: Sample Size Calculation (cont’d)
Sample size calculation for a continuous endpoint
Fixed sample analysis, one-sided significance level 2.5%, power 80%. The results were calculated for a two-sample t-test, H0: mu(1) - mu(2) = 0, H1: effect = 12, standard deviation = 15.
| Stage | Fixed |
|---|---|
| Stage level (one-sided) | 0.0250 |
| Efficacy boundary (z-value scale) | 1.960 |
| Efficacy boundary (t) | 8.438 |
| Number of subjects | 51.0 |
Legend:
- (t): treatment effect scale
Step 1: Sample Size Calculation (cont’d)
Using fetch() we can extract specific quantities from the result object.
Step 2: Calculating Power
Now let’s go the other way around - calculate the power for given sample size, and vary other parameters. This helps us to:
- Verify the Assumptions: Confirm that the chosen sample size yields the expected 80% power.
- Explore Outcome Variability: Observing power across different situations can reveal potential inconsistencies in achieving the target power, especially with variations in standard deviation or effect size.
- Adjust Study Design: If the results show that power is consistently below the target, we might need to increase the sample size or reconsider assumptions.
Step 2: Calculating Power (cont’d)
Using the rpact package, we can calculate the power of the study under the defined parameters. Here’s how:
# Define parameters based on initial assumptions
calculatedSampleSize <- ceiling(sampleSizeResult$numberOfSubjects)
powerResult <- design |>
getPowerMeans(
groups = 2, # Two groups: Treatment and Placebo
alternative = 12, # Expected effect size
stDev = 15, # Common standard deviation
maxNumberOfSubjects = calculatedSampleSize
)Step 2: Calculating Power (cont’d)
Power calculation for a continuous endpoint
Fixed sample analysis, one-sided significance level 2.5%. The results were calculated for a two-sample t-test, H0: mu(1) - mu(2) = 0, power directed towards larger values, H1: effect = 12, standard deviation = 15, number of subjects = 52.
| Stage | Fixed |
|---|---|
| Stage level (one-sided) | 0.0250 |
| Efficacy boundary (z-value scale) | 1.960 |
| Efficacy boundary (t) | 8.356 |
| Power | 0.8075 |
| Number of subjects | 52.0 |
Legend:
- (t): treatment effect scale
Step 2: Calculating Power (cont’d)
- The results show that in 80.75% of the trials the test successfully detects a true difference between treatment and control group regarding change from baseline.
- Next steps: proceed with further setup and explore possible adjustments based on the outcomes.
Exploring Possible Adjustments Based on Power Calculation Outcomes
- Varying Effect Size: If the expected effect size of 12 mmHg is optimistic, we could calculate a scenario with a smaller effect size (e.g., 10 mmHg) to see how it affects the required sample size and power.
- Varying Standard Deviation: The assumed standard deviation of 15 mmHg might vary across patient groups. Increasing it to 16 or 17 mmHg would allow us to explore how higher variability impacts power and the required sample size.
Possible Adjustments
Here’s R code to analyze the scenarios described above:
# Define scenarios for adjustments
scenarios <- list(
list(alternative = 10, stDev = 15), # 1: Reduced effect
list(alternative = 10, stDev = 14), # 2: Reduced effect + sd
list(alternative = 11, stDev = 15), # 3: Reduced effect
list(alternative = 11, stDev = 14), # 4: Reduced effect + sd
list(alternative = 12, stDev = 15), # 5: Base scenario
list(alternative = 13, stDev = 16), # 6: Incr. sd + effect
list(alternative = 13, stDev = 17), # 7: Incr. sd + effect
list(alternative = 12, stDev = 16), # 8: Increased sd
list(alternative = 12, stDev = 17) # 9: Increased sd
)
# Run calculations for each scenario
results <- scenarios |>
lapply(function(scenario) {
getPowerMeans(
design = design,
groups = 2,
alternative = scenario$alternative,
stDev = scenario$stDev,
maxNumberOfSubjects = 52
)
})
# Fetch only the power from the result objects
x <- sapply(results, function(result) {
result |> fetch("Overall reject")
})Possible Adjustments (cont’d)
| alternative | stDev | power | |
|---|---|---|---|
| Scenario 1 | 10 | 15 | 0.6544534 |
| Scenario 2 | 10 | 14 | 0.7142007 |
| Scenario 3 | 11 | 15 | 0.7366421 |
| Scenario 4 | 11 | 14 | 0.7933714 |
| Scenario 5 | 12 | 15 | 0.8074858 |
| Scenario 6 | 13 | 16 | 0.8193392 |
| Scenario 7 | 13 | 17 | 0.7715373 |
| Scenario 8 | 12 | 16 | 0.7555122 |
| Scenario 9 | 12 | 17 | 0.7040188 |
Possible Adjustments (cont’d)
Interpreting the results from the different scenarios:
- Base scenario (52 subjects, 12 mmHg effect, 15 mmHg SD): Verifying the original power (around 80.75%).
- Reduced effect size (10 mmHg effect): With a smaller effect size, the power drops to 65.45%.
- Increased standard deviation (17 mmHg SD): Increasing variability lowers the power to 70.4%, as greater variability results in less precise outcomes.
By exploring these adjustments, we gain insight into whether a larger sample or refined target parameters might be needed for a more reliable assessment of the new treatments’s efficacy.
Exploring Scenarios with Larger Treatment Group
- Explore scenarios with unequal allocation ratios, assigning more participants to the treatment group than to the control group.
- This approach allows more patients the potential benefit of the new therapy while still maintaining a control group for reliable statistical comparison.
- The
allocationRatioPlannedargument inrpactenables us to specify these ratios.
Scenario Adjustments Using Unequal Allocation Ratios
Here are the new scenarios we will examine:
- Base scenario (1:1 allocation): Both groups (treatment and control) have equal numbers of participants.
- 2:1 allocation: For every 1 participant in the control group, 2 are assigned to the treatment group.
- 3:1 allocation: For every 1 participant in the control group, 3 are assigned to the treatment group.
Comparing Different Allocation Ratios
Using the rpact package, we can set up and calculate these scenarios with the allocationRatioPlanned parameter:
# Define the initial design
design <- getDesignGroupSequential(
kMax = 1,
alpha = 0.025,
beta = 0.2,
sided = 1
)
# Define scenarios for different allocation ratios
scenarios <- list(
list(allocationRatio = 1), # 1:1 allocation
list(allocationRatio = 2), # 2:1 allocation
list(allocationRatio = 3) # 3:1 allocation
)
# Run calculations for each scenario with specified allocation ratios
results <- scenarios |>
lapply(function(scenario) {
getPowerMeans(
design = design,
groups = 2,
alternative = 12,
stDev = 15,
maxNumberOfSubjects = 50,
allocationRatioPlanned = scenario$allocationRatio
)
})
# Fetch only the power from the result objects
x <- sapply(results, function(result) {
result |> fetch("Overall reject")
})Comparing Different Allocation Ratios (cont’d)
| allocationRatio | power | |
|---|---|---|
| Scenario 1 | 1 | 0.7914502 |
| Scenario 2 | 2 | 0.7431284 |
| Scenario 3 | 3 | 0.6701351 |
Let’s assume that a 2:1 allocation shall be used going forward.
Continuing with a Group-Sequential Design to Open Up the Opportunity for Early Stopping
Given the slight reduction in power observed with the 2:1 allocation ratio (Power = 74.3%), we can implement a group-sequential design with an interim analysis instead of simply increasing the sample size.
- Planned Interim Analysis:
- After enrolling a number yet to be determined participants, an interim analysis will be conducted.
- Possible Outcomes:
- If the treatment shows significant efficacy at the interim point, the study can be stopped early to avoid additional recruitment.
- If efficacy is not yet demonstrated, we proceed to the full planned sample size.
Step 1: Calculate the sample size for a fixed design
First we start again with a fixed sample size design:
nFixed
58 We need numberOfSubjectsFixed for the subsequent power analysis.
Step 2: Define the Group-Sequential Design with an Interim Analysis
- We’ll use
rpactto define a group-sequential design with two planned analyses. - The first analysis will occur after enrolling
nsubjects, with a final analysis if the study proceeds to the full sample. nshall be determined by exploring different information rates
Design Comparison in R
Here’s how to set up the design comparison in R:
First we define the possible information rates at which to conduct the interim analysis.
Design Comparison in R (cont’d)
# Run calculations for each scenario
results <- scenarios |>
lapply(function(scenario) {
getDesignGroupSequential(
informationRates = c(scenario$informationRate, 1),
alpha = 0.025,
sided = 1,
typeOfDesign = "OF"
) |>
getPowerMeans(
groups = 2,
alternative = 12,
stDev = 15,
maxNumberOfSubjects = numberOfSubjectsFixed, # 58
allocationRatioPlanned = 2
)
})Design Comparison in R (cont’d)
| informationRate | expectedNumberOfSubjects | earlyStop | |
|---|---|---|---|
| Scenario 1 | 0.5 | 51.86201 | 0.2116549 |
| Scenario 2 | 0.6 | 49.88180 | 0.3499223 |
| Scenario 3 | 0.7 | 49.56749 | 0.4846273 |
| Scenario 4 | 0.8 | 50.99013 | 0.6042989 |
| Scenario 5 | 0.9 | 53.90422 | 0.7061693 |
In this example we decide to use information rate 0.7 because the expected number of subjects is lowest and the probability for an early stopping is nearly 50%.
Sample Size with an Interim Analysis
design <- getDesignGroupSequential(
informationRates = c(0.7, 1),
alpha = 0.025, # Overall significance level
beta = 0.2, # Power 80%
sided = 1, # One-sided test
typeOfDesign = "OF" # O'Brien & Fleming design
)
sampleSizeResult <- getSampleSizeMeans(
design = design,
groups = 2,
alternative = 12, # Target effect
stDev = 15, # Common standard deviation
allocationRatioPlanned = 2 # 2:1 allocation
)
# Print the summary of the results
sampleSizeResult |>
summary()Sample size calculation for a continuous endpoint
Sequential analysis with a maximum of 2 looks (group sequential design), one-sided overall significance level 2.5%, power 80%. The results were calculated for a two-sample t-test, H0: mu(1) - mu(2) = 0, H1: effect = 12, standard deviation = 15, planned allocation ratio = 2.
| Stage | 1 | 2 |
|---|---|---|
| Planned information rate | 70% | 100% |
| Cumulative alpha spent | 0.0082 | 0.0250 |
| Stage levels (one-sided) | 0.0082 | 0.0223 |
| Efficacy boundary (z-value scale) | 2.400 | 2.008 |
| Efficacy boundary (t) | 12.508 | 8.566 |
| Cumulative power | 0.4861 | 0.8000 |
| Number of subjects | 40.7 | 58.2 |
| Expected number of subjects under H1 | 49.7 | |
| Exit probability for efficacy (under H0) | 0.0082 | |
| Exit probability for efficacy (under H1) | 0.4861 |
Legend:
- (t): treatment effect scale
Sample Size with an Interim Analysis (cont’d)
Step 3: Calculate the Power of the Group-Sequential Design
To confirm that the design meets the target power while allowing for early stopping, we calculate the power of the study under the adjusted parameters:
powerResult <- getPowerMeans(
design = design,
groups = 2,
alternative = 12, # Expected effect size
stDev = 15, # Common standard deviation
# Sample size per stage from previous calculation
maxNumberOfSubjects = ceiling(sampleSizeResult$numberOfSubjects)[2, 1],
allocationRatioPlanned = 2 # Allocation ratio 2:1
)
# Print the summary of the results
powerResult |>
summary()Power calculation for a continuous endpoint
Sequential analysis with a maximum of 2 looks (group sequential design), one-sided overall significance level 2.5%. The results were calculated for a two-sample t-test, H0: mu(1) - mu(2) = 0, power directed towards larger values, H1: effect = 12, standard deviation = 15, maximum number of subjects = 59, planned allocation ratio = 2.
| Stage | 1 | 2 |
|---|---|---|
| Planned information rate | 70% | 100% |
| Cumulative alpha spent | 0.0082 | 0.0250 |
| Stage levels (one-sided) | 0.0082 | 0.0223 |
| Efficacy boundary (z-value scale) | 2.400 | 2.008 |
| Efficacy boundary (t) | 12.416 | 8.506 |
| Cumulative power | 0.4930 | 0.8057 |
| Number of subjects | 41.3 | 59.0 |
| Expected number of subjects under H1 | 50.3 | |
| Exit probability for efficacy (under H0) | 0.0082 | |
| Exit probability for efficacy (under H1) | 0.4930 |
Legend:
- (t): treatment effect scale
Adding a Futility Analysis
- Now we want to find a futility bound for the interim analysis that depends on a clinical relevant threshold.
- We assume that it makes no sense to continue the analysis when the observed treatment effect is 5 mmHg or less
- The minimum futility bound must be 0 because otherwise the effect would go in the wrong direction
- Ultimately, we go for a boundary of 2 mmHg on treatment effect scale
Adding a Futility Analysis (cont’d)
# Run calculations for each scenario
results <- scenarios |>
lapply(function(scenario) {
getDesignGroupSequential(
informationRates = c(0.7, 1),
futilityBounds = scenario$futilityBounds,
alpha = 0.025,
sided = 1,
typeOfDesign = "OF"
) |>
getSampleSizeMeans(
groups = 2,
alternative = 12,
stDev = 15,
allocationRatioPlanned = 2
)
})Adding a Futility Analysis (cont’d)
# Display results for each scenario
x <- sapply(results, function(result) {
result |> fetch("Futility bounds (treatment effect scale)")
})
#| echo: true
scenarios |>
bind_rows() |>
mutate("Futility bounds (treatment effect scale)" = x) |>
as.data.frame() |>
set_rownames(paste("Scenario", 1:length(x))) |>
kable()| futilityBounds | Futility bounds (treatment effect scale) | |
|---|---|---|
| Scenario 1 | -1.0 | -5.050045 |
| Scenario 2 | -0.5 | -2.512664 |
| Scenario 3 | 0.0 | 0 |
| Scenario 4 | 0.5 | 2.508815 |
| Scenario 5 | 1.0 | 4.97699 |
| Scenario 6 | 1.5 | 7.102175 |
Find Futility Bound on the Z Scale
We will first search the futility bound on the Z scale that corresponds to a treatment effect of 2 mmHg. We do this with stats::uniroot():
One Dimensional Root (Zero) Finding
Description
The function uniroot searches the interval from lower to upper for a root (i.e., zero) of the function f with respect to its first argument.
Find Futility Bound on the Z Scale (cont’d)
soughtBoundaryTreatmentEffectScale <- 2
futilityBound <- uniroot(
function(x) {
soughtBoundaryTreatmentEffectScale -
getDesignGroupSequential(
informationRates = c(0.7, 1),
futilityBounds = x,
alpha = 0.025,
sided = 1,
typeOfDesign = "OF"
) |>
getSampleSizeMeans(
groups = 2,
alternative = 12,
stDev = 15,
allocationRatioPlanned = 2
) |>
fetch("Futility bounds (treatment effect scale)") |>
as.numeric()
},
lower = 0,
upper = 2
)$root
futilityBound[1] 0.3985836Find Futility Bound on the Z Scale (cont’d)
Alternatively, we can also calculate this in an approximate closed form here via:
\[ u_{1}^{0} = \delta_{0} \sqrt{\frac{n_{11}n_{21}}{n_{11} + n_{21}}} / \sigma \]
Sample Size with Futility Bound
Sample Size with Futility Bound (cont’d)
Sample size calculation for a continuous endpoint
Sequential analysis with a maximum of 2 looks (group sequential design), one-sided overall significance level 2.5%, power 80%. The results were calculated for a two-sample t-test, H0: mu(1) - mu(2) = 0, H1: effect = 12, standard deviation = 15, planned allocation ratio = 2.
| Stage | 1 | 2 |
|---|---|---|
| Planned information rate | 70% | 100% |
| Cumulative alpha spent | 0.0082 | 0.0250 |
| Stage levels (one-sided) | 0.0082 | 0.0223 |
| Efficacy boundary (z-value scale) | 2.400 | 2.008 |
| Futility boundary (z-value scale) | 0.401 | |
| Efficacy boundary (t) | 12.496 | 8.558 |
| Futility boundary (t) | 2.013 | |
| Cumulative power | 0.4869 | 0.8000 |
| Number of subjects | 40.8 | 58.3 |
| Expected number of subjects under H1 | 49.4 | |
| Overall exit probability (under H0) | 0.6641 | |
| Overall exit probability (under H1) | 0.5116 | |
| Exit probability for efficacy (under H0) | 0.0082 | |
| Exit probability for efficacy (under H1) | 0.4869 | |
| Exit probability for futility (under H0) | 0.6559 | |
| Exit probability for futility (under H1) | 0.0246 |
Legend:
- (t): treatment effect scale
Plot the Design with Futility Bound
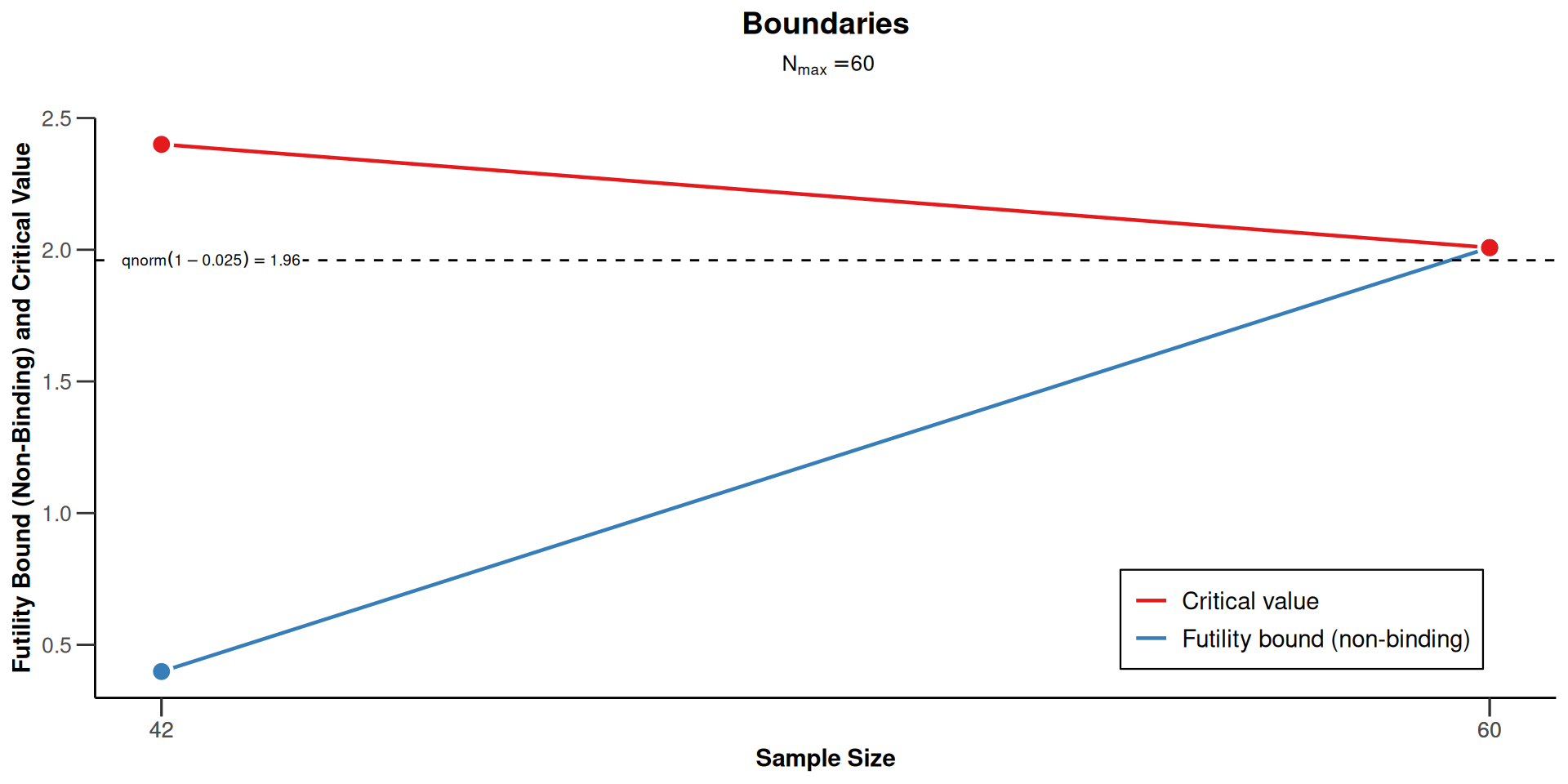
Plot the Design with Futility Bound (cont’d)
Plot the Design with Futility Bound (cont’d)
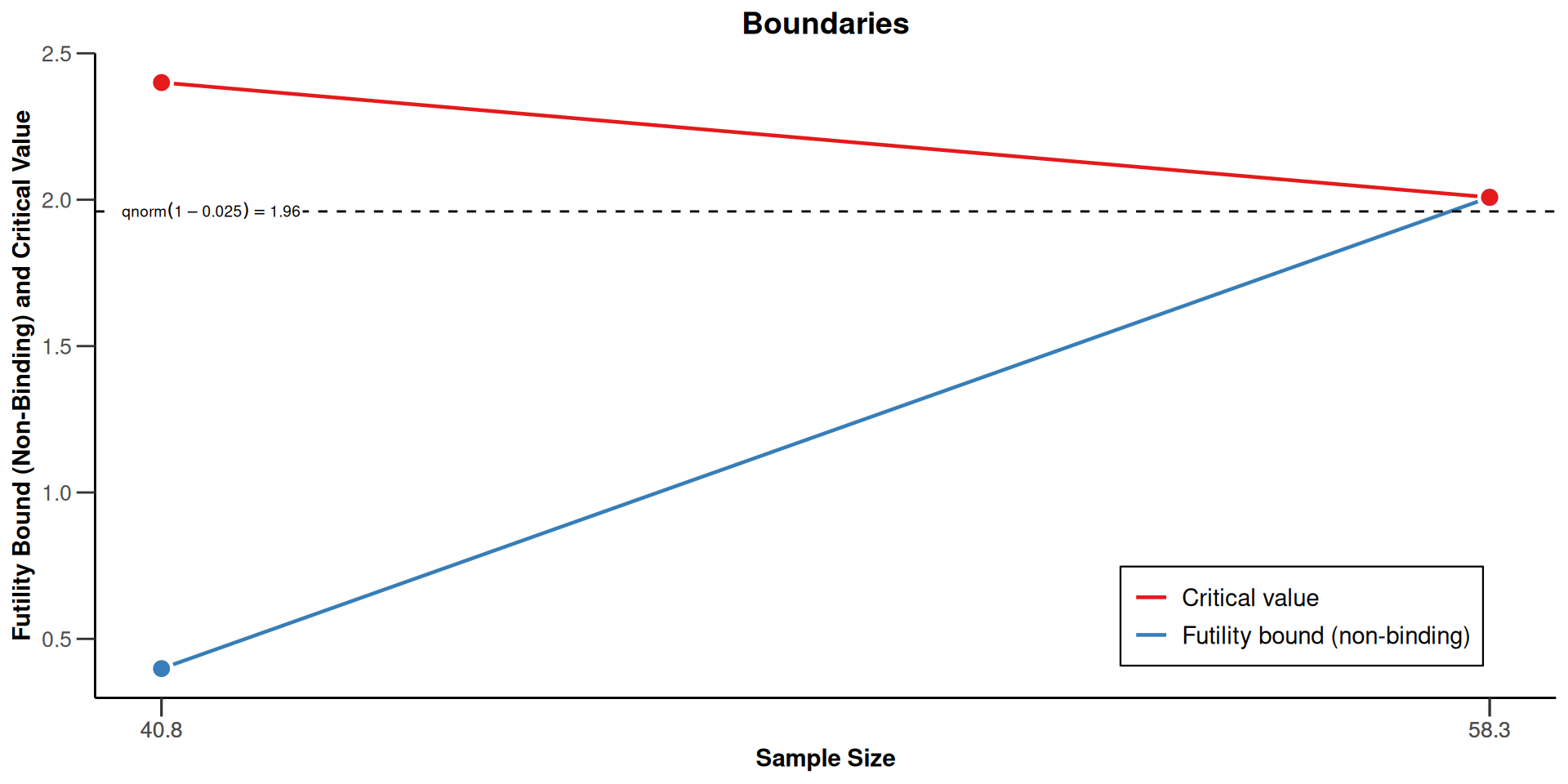
Simulations to Study Operating Characteristics
In this example, we could of course skip simulations, because for a continuous outcome with known standard deviation we can calculate everything in closed form.
However, to illustrate the simulation capabilities of
rpact, we perform a simulation study to confirm the operating characteristics of our group-sequential design with futility stopping.This is important for non-normal endpoints and adaptive designs where closed form solutions are not available.
Simulation Setup
Now we just call getSimulationMeans() with the design and parameters defined above:
simResults <- getDesignGroupSequential(
informationRates = c(0.7, 1),
futilityBounds = futilityBound,
alpha = 0.025,
sided = 1,
typeOfDesign = "OF"
) |>
getSimulationMeans(
groups = 2,
alternative = c(0, 12), # H0 and H1
stDev = 15,
allocationRatioPlanned = 2,
plannedSubjects = c(41, 59),
maxNumberOfIterations = 1000,
seed = 12345
)Simulation Results (cont’d)
Simulation of a continuous endpoint
Sequential analysis with a maximum of 2 looks (group sequential design), one-sided overall significance level 2.5%. The results were simulated for a two-sample t-test (normal approximation), H0: mu(1) - mu(2) = 0, power directed towards larger values, H1: effect as specified, standard deviation = 15, planned cumulative sample size = c(41, 59), planned allocation ratio = 2, simulation runs = 1000, seed = 12345.
| Stage | 1 | 2 |
|---|---|---|
| Planned information rate | 70% | 100% |
| Cumulative alpha spent | 0.0082 | 0.0250 |
| Stage levels (one-sided) | 0.0082 | 0.0223 |
| Efficacy boundary (z-value scale) | 2.400 | 2.008 |
| Futility boundary (z-value scale) | 0.399 | |
| Cumulative power, alt. = 0 | 0.0040 | 0.0210 |
| Cumulative power, alt. = 12 | 0.5060 | 0.8120 |
| Stage-wise number of subjects, alt. = 0 | 41.0 | 18.0 |
| Stage-wise number of subjects, alt. = 12 | 41.0 | 18.0 |
| Expected number of subjects under H1, alt. = 0 | 47.4 | |
| Expected number of subjects under H1, alt. = 12 | 49.5 | |
| Conditional power (achieved), alt. = 0 | 0.1604 | |
| Conditional power (achieved), alt. = 12 | 0.5304 | |
| Exit probability for efficacy, alt. = 0 | 0.0040 | |
| Exit probability for efficacy, alt. = 12 | 0.5060 | |
| Exit probability for futility, alt. = 0 | 0.6390 | |
| Exit probability for futility, alt. = 12 | 0.0230 |
Legend:
- alt.: alternative
Simulation Results (cont’d)
Interpretation:
- We can see the type I error control:
Cumulative power, alt. = 0is 2.1% at the final analysis. - We can see the power:
Cumulative power, alt. = 12is 81.2% at the final analysis. - We can confirm that around half of the trials would already stop successfully at the interim analysis under the alternative hypothesis:
Exit probability for efficacy, alt. = 12is 50.6%
Collect and Analyse Data
Download the example dataset: trial_data_stage1.csv
# A tibble: 42 × 4
subjectId group bloodPressureBaseline bloodPressure
<int> <fct> <dbl> <dbl>
1 1 1 150. 138.
2 2 1 144. 160.
3 3 1 151. 128.
4 4 1 157. 147.
5 5 1 153. 136.
6 6 1 155. 146.
7 7 1 154. 120.
8 8 1 150. 173.
9 9 1 149. 147.
10 10 1 145. 113.
# ℹ 32 more rowsThe Dataset
# A tibble: 42 × 5
subjectId group bloodPressureBaseline bloodPressure bloodPressureDiff
<int> <fct> <dbl> <dbl> <dbl>
1 1 1 150. 138. -11.6
2 2 1 144. 160. 15.8
3 3 1 151. 128. -23.2
4 4 1 157. 147. -9.72
5 5 1 153. 136. -17.5
6 6 1 155. 146. -8.76
7 7 1 154. 120. -34.4
8 8 1 150. 173. 22.9
9 9 1 149. 147. -2.66
10 10 1 145. 113. -31.7
# ℹ 32 more rowsData Analysis
rpact requires the summary statistics, therefore let’s calculate them:
dataSummary <- trialData |>
group_by(group) |>
summarise(
n = n(),
meanBaseline = mean(bloodPressureBaseline),
stDevBaseline = sd(bloodPressureBaseline),
meanTherapy = mean(bloodPressure),
stDevTherapy = sd(bloodPressure),
mean = mean(bloodPressureDiff),
stDev = sd(bloodPressureDiff)
)
dataSummary |>
kable()| group | n | meanBaseline | stDevBaseline | meanTherapy | stDevTherapy | mean | stDev |
|---|---|---|---|---|---|---|---|
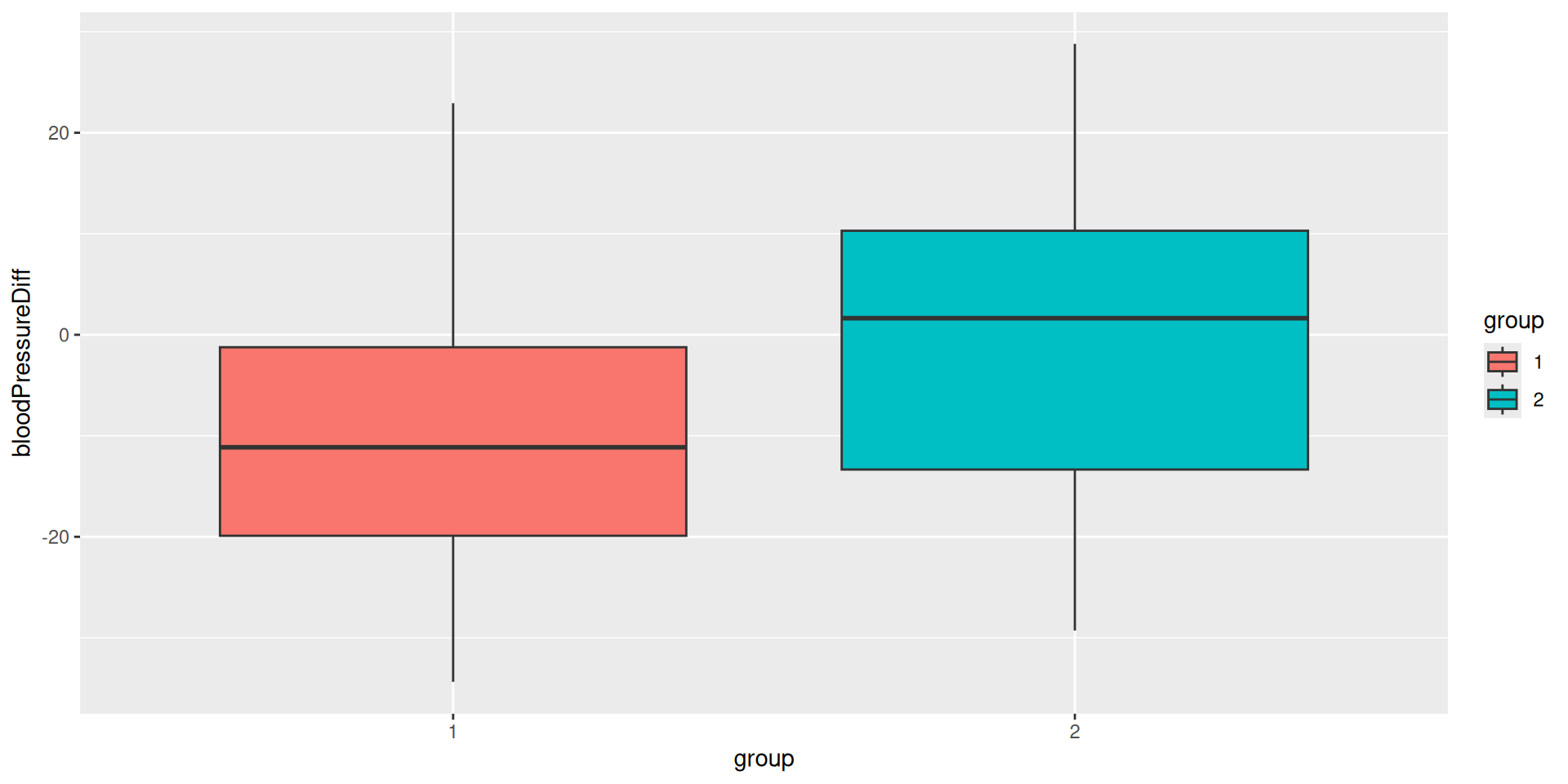
| 1 | 28 | 149.2407 | 5.677756 | 139.0127 | 14.33155 | -10.2280732 | 14.07891 |
| 2 | 14 | 148.1788 | 3.743160 | 147.7018 | 17.22372 | -0.4770206 | 17.14525 |
Plot the trial data
Create an rpact Dataset
Create an rpact dataset from summary statistics of the trial data:
Interim Analysis: First Look Results
Analysis results for a continuous endpoint
Sequential analysis with 2 looks (group sequential design), one-sided overall significance level 2.5%. The results were calculated using a two-sample t-test, equal variances option. H0: mu(1) - mu(2) = 0 against H1: mu(1) - mu(2) < 0. The conditional power calculation with planned sample size is based on planned allocation ratio = 2, overall effect = -9.751, and assumed standard deviation = 15.
| Stage | 1 | 2 |
|---|---|---|
| Planned information rate | 70% | 100% |
| Cumulative alpha spent | 0.0082 | 0.0250 |
| Stage levels (one-sided) | 0.0082 | 0.0223 |
| Efficacy boundary (z-value scale) | 2.400 | 2.008 |
| Cumulative effect size | -9.751 | |
| Cumulative (pooled) standard deviation | 15.144 | |
| Overall test statistic | -1.967 | |
| Overall p-value | 0.0281 | |
| Test action | continue | |
| Conditional rejection probability | 0.2271 | |
| Planned sample size | 18 | |
| Conditional power | 0.7094 | |
| 95% repeated confidence interval | [-22.171; 2.669] | |
| Repeated p-value | 0.0636 |
Note that we must set directionUpper = FALSE because we have a higher blood pressure reduction in the treatment group, i.e., the blood pressure reduction difference between control and treatment is negative.
Final analysis: Second Look Results
Download the example dataset: trial_data.csv
# A tibble: 60 × 5
subjectId stage group bloodPressureBaseline bloodPressure
<int> <int> <fct> <dbl> <dbl>
1 1 1 1 150. 138.
2 2 1 1 144. 160.
3 3 1 1 151. 128.
4 4 1 1 157. 147.
5 5 1 1 153. 136.
6 6 1 1 155. 146.
7 7 1 1 154. 120.
8 8 1 1 150. 173.
9 9 1 1 149. 147.
10 10 1 1 145. 113.
# ℹ 50 more rowsAlternative Calculation
Alternatively we can use the emmeans package together with a model definition to load the raw data into an rpact dataset.
library(emmeans)
trialData$group <- relevel(trialData$group, ref = "2")
trialData$bloodPressureDiff <-
trialData$bloodPressure - trialData$bloodPressureBaseline
dataset <- getDataset(
lm(bloodPressureDiff ~ group, data = trialData, subset = (stage == 1)) |>
emmeans("group"),
lm(bloodPressureDiff ~ group, data = trialData, subset = (stage == 2)) |>
emmeans("group")
)Note: Here we are not interested in the model output but we need it to compute the estimated marginal means (EMMs; least-squares means) for the factor group in the model. This type of raw data import is particularly useful when the raw data contains covariates that need to be adjusted for.
Alternative Calculation (cont’d)
Dataset of means
The dataset contains the sample sizes, means, and standard deviations of one treatment and one control group. The total number of looks is two; stage-wise and cumulative data are included.
| Stage | 1 | 1 | 2 | 2 |
|---|---|---|---|---|
| Group | 1 | 2 | 1 | 2 |
| Stage-wise sample size | 28 | 14 | 12 | 6 |
| Cumulative sample size | 28 | 14 | 40 | 20 |
| Stage-wise mean | -10.228 | -0.477 | -13.268 | 0.313 |
| Cumulative mean | -10.228 | -0.477 | -11.140 | -0.240 |
| Stage-wise standard deviation | 15.144 | 15.144 | 15.275 | 15.275 |
| Cumulative standard deviation | 15.144 | 15.144 | 15.052 | 14.780 |
Alternative Calculation (cont’d)
Note that the standard deviation looks different depending of the method you use to import the raw data:
- dplyr: group-wise standard deviations
- emmeans: pooled standard deviations
For the analysis with rpact this makes no difference.
Alternative Calculation (cont’d)
Analysis results for a continuous endpoint
Sequential analysis with 2 looks (group sequential design), one-sided overall significance level 2.5%. The results were calculated using a two-sample t-test, equal variances option. H0: mu(1) - mu(2) = 0 against H1: mu(1) - mu(2) < 0.
| Stage | 1 | 2 |
|---|---|---|
| Planned information rate | 70% | 100% |
| Cumulative alpha spent | 0.0082 | 0.0250 |
| Stage levels (one-sided) | 0.0082 | 0.0223 |
| Efficacy boundary (z-value scale) | 2.400 | 2.008 |
| Cumulative effect size | -9.751 | -10.900 |
| Cumulative (pooled) standard deviation | 15.144 | 14.964 |
| Overall test statistic | -1.967 | -2.660 |
| Overall p-value | 0.0281 | 0.0050 |
| Test action | continue | reject |
| Conditional rejection probability | 0.2271 | |
| 95% repeated confidence interval | [-22.171; 2.669] | [-19.311; -2.489] |
| Repeated p-value | 0.0636 | 0.0054 |
| Final p-value | 0.0107 | |
| Final confidence interval | [-18.228; -1.525] | |
| Median unbiased estimate | -10.004 |
Thank you!
Questions and Answers

